current research directions
cell biology of meiotic drive: genetic cheating in violation of Mendel’s First Law
Every high school biology student learns about the 3:1 phenotype ratio observed by Mendel in his monohybrid crosses of pea plants. Underlying this observation, and at the core of Mendelian genetics, is the concept that gametes are equally likely to carry either allele of a gene. Selfish genetic elements can cheat by violating this principle to increase their representation in the gametes (and therefore in the progeny) by meiotic drive. Examples from a range of eukaryotes show that meiotic drive is a widespread phenomenon that impacts many aspects of reproduction, genetics, and evolution (Werren 2011, Rice 2013, Helleu et al. 2015, Lindholm et al. 2016). These violations of Mendel’s First Law should perhaps not surprise us because the opportunity to cheat is inherent in the haploid-diploid life cycle that underlies sexual reproduction in all eukaryotes, with genetic elements competing for transmission to the offspring through meiosis, the process by which haploids are generated. Female meiosis provides the most straightforward way to cheat because only one egg is produced from two rounds of cell division, so selfish elements compete for transmission to the egg (Sandler and Novitski 1957, Pardo-Manuel de Villena and Sapienza, 2001). Our studies focus on the cell biology of female meiotic drive, using a mouse model system in which we can directly observe drive and ask mechanistic questions (Chmátal et al. 2014, Iwata-Otsubo et al. 2017, Akera et al. 2017, Akera et al. 2019, Kumon et al. 2021). The area is wide open, and the significance of meiotic drive for genetics and evolutionary and reproductive biology makes the cell biological questions particularly exciting. Furthermore, understanding how selfish centromeres cheat the meiotic segregation machinery will provide new insights into centromere biology and meiotic cell division. Read more or see our papers in this area …
challenges of the extended meiotic cell cycle in mammalian oocytes and germline centromere inheritance
The extended prophase I arrest (lasting months to decades) in mammalian oocytes raises questions about how chromosome cohesion and centromere identity are maintained. Cohesin proteins and CENP-A nucleosomes, which define the centromere, assemble onto chromosomes before the prophase arrest and must persist through the arrest. Using mouse oocytes as our experimental system, we showed that cohesin proteins are gradually lost from chromosomes during prophase I and that the resulting cohesion defects can explain most of the age-related chromosome segregation errors (Duncan et al. 2009, Chiang et al. 2010, 2011). Our findings established a mechanistic basis for the well-known increase in aneuploidy associated with advanced maternal age. In contrast, we showed that CENP-A nucleosomes are stable for the fertile lifespan of the female (>1 yr) in the absence of any new assembly (Smoak et al. 2016). This remarkable stability underlies centromere inheritance through the female germline. After fertilization, we showed that new centromere chromatin assembly depends on centromere DNA sequences and on the maternal Cenpa genotype in addition to the established epigenetic pathway (Das et al. 2022). See our papers in this area …
 |
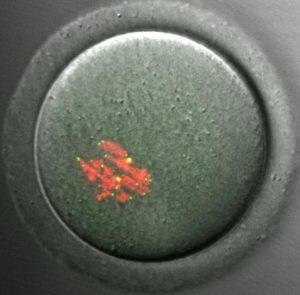
CENP-A staining in a mouse oocyte at metaphase I. CENP-A persists at centromeres for >1 year after the gene has been deleted in the oocyte, demonstrating the remarkable stability of centromeric nucleosomes.
See Smoak et al. 2016. |
chemical optogenetic tools to manipulate living cells
Tools for manipulating protein levels at intracellular structures, with temporal and spatial control, will enable the next generation of experiments to dissect dynamic systems in cell biology. Kinetochores are a prime example: multi-subunit complexes whose function depends on dynamic changes in localization of kinases, MT-associated proteins, and checkpoint signaling proteins throughout cell division. We developed optogenetic probes, in collaboration with the Chenoweth Lab, based on photo-caged or photo-cleavable chemical dimerizers. These probes allow us to recruit proteins to individual kinetochores or other intracellular structures with light, with both spatial and temporal precision. We have applied these tools to manipulate checkpoint signaling, molecular motors, and Aurora kinase activity at kinetochores (Ballister et al. 2014, Zhang et al. 2017, Aonbangkhen et al. 2018, Chen et al. 2021), and nuclear body phase separation at telomeres (Zhang et al. 2020). Read more or see our papers in this area …
regulation of kinetochore-microtubule interactions
How MT attachments are regulated to ensure accurate chromosome segregation is a long-standing question in cell biology. We addressed this question at multiple levels, using two different systems. In cultured somatic cells, we established a mechanism for phosphatase recruitment to kinetochores to oppose Aurora B (Liu et al. 2010) and showed that Polo-like kinase-1 (Plk1) stabilizes kinetochore MTs in opposition to Aurora B (Liu et al. 2012). Using mouse oocytes as a meiotic model system, we showed that gradually increasing Cdk1 activity acts as a timing mechanism for stabilizing MT attachments after spindle formation (Davydenko et al. 2013). We also demonstrated spatial regulation of attachments by Aurora A at spindle poles (Chmatal et al. 2015), complementary to tension-dependent regulation by Aurora B at centromeres. Most recently, we used an optogenetic approach to manipulate Aurora B at individual kinetochores and showed that the response to kinase activity depends on centromere tension to induce distinct error correction mechanisms (Chen et al. 2021). See our papers in this area …
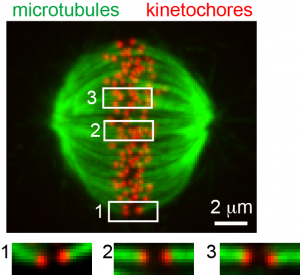 |
Kinetochore microtubule bi-orientation. A metaphase cell stained for microtubules (green) and kinetochores (red). The image is a maximal intensity projection of a confocal z-series. Insets are optical sections showing bi-oriented sister kinetochore pairs.
From Liu et al. 2012. |
previous research areas
spatial patterns of Aurora B kinase activity
Aurora B is a key mitotic kinase that redistributes from centromeres to the spindle midzone at anaphase onset, as part of the chromosome passenger complex. To determine how spatial changes in Aurora B signaling regulate cell division, we developed FRET-based biosensors to track changes in kinase activity in living cells with high temporal and spatial resolution. We measured spatial phosphorylation gradients around the spindle midzone in anaphase and around centromeres before anaphase onset, and we established a model for how spatially regulated Aurora B kinase activity can selectively destabilize kinetochore-MT attachment errors (Fuller et al. 2008, Liu et al. 2009, Wang et al. 2011, Zaytsev et al. 2016). Our model is widely cited and is included in a recent textbook (Lodish et al. Molecular Cell Biology, 7th Edition, Figure 19-25). See our papers in this area …
collaborators
Ben Black, Department of Biochemistry and Biophysics
Dennis Discher, Department of Chemical and Biomolecular Engineering
Ekaterina Grishchuk, Department of Physiology
Andrea Liu, Department of Physics and Astronomy
Dave Chenoweth, Department of Chemistry
Roger Greenberg, Department of Cancer Biology
Mia Levine, Department of Biology
Richard Schultz, Department of Biology
Iain Cheeseman, Whitehead Institute and MIT
Alexey Khodjakov, Wadsworth Center
research support (past and present)
National Institute of General Medical Sciences (NIGMS)
National Human Genome Research Institute (NHGRI)
National Institute of Child Health and Human Development (NICHD)
Searle Scholars Award
Discovering the Future Research Grant, University of Pennsylvania
Abramson Cancer Center, University of Pennsylvania
Physical Sciences Oncology Center at Penn (PSOC)
Penn Center for Genome Integrity
Penn Center for Molecular Studies in Digestive and Liver Diseases
Emerson Collective Cancer Research Fund
Nano/Bio Interface Center, University of Pennsylvania
University Research Foundation, University of Pennsylvania
Institute on Aging, University of Pennsylvania
Penn Genome Frontiers Institute
Translational Biomedical Imaging Center, Institute for Translational Medicine and Therapeutics, University of Pennsylvania
Penn Center for Study of Epigenetics in Reproduction