Dr. Lucy Shan successfully defended her Ph.D. on Wednesday, November 25 virtually via zoom. Her thesis project identified possible novel roles of RNA binding proteins involved in blood cell development and in the development of moss using high throughput sequencing and proteomic data. Congratulations Dr. Shan!
Updated Publications List
Sorry for the long delay!! The Publications list of the Gregory Lab has now been updated to encompass all of the most recent articles, reviews, and book chapters from the lab!!
Our new Review Article available now!
Our new Review Article describing the recent analyses on RNA secondary structure and RBP-RNA interactions in Arabidopsis is now published in WIREs RNA!! Congrats to Marianne and Shawn on a really nice article. This article can be found at the following link.
This article was also highlighted, and the highlight document can be found at the following link for those interested!
http://www.advancedsciencenews.com/rna-structure-binding-coordination-arabidopsis
Manuscript describing our genome-wide Topoisomerase cleavage assay published
Our collaborative study where we and the Felix lab developed a high-throughput sequencing-based approach for mapping TOP2A cleavage on a genome-wide scale is now online at Genome Research. This was a true labor of love to get this study to completion, but I could not be happier to have this assay finally published!
http://genome.cshlp.org/content/early/2017/04/06/gr.211615.116.long
Congrats to Xiang and James for getting this study passed the finish line!
Study identifying novel post-transcriptional regulators of root hair cell fate published
Our study identifying novel post-transcriptional regulators of root hair cell fate is now published in Developmental Cell! We also are the cover article for this issue!
http://www.cell.com/developmental-cell/fulltext/S1534-5807%2817%2930202-2
Congratulations, Shawn on a great study. Also, a huge thank you to our fantastic collaborative team.
A press release about this work can be found through the following link.
https://news.upenn.edu/news/penn-team-identifies-genetic-target-growing-hardier-plants-under-stress
Plant co-translational RNA decay manuscript online now
Our manuscript that describes some of the features and factors involved in plant co-translational RNA decay is being published at Plant Cell. An online early version of our manuscript can be found at the following link.
http://www.plantcell.org/content/early/2016/10/07/tpc.16.00456.abstract
Congrats to postdoc Xiang Yu and former postdoc Matthew Willmann (now head of the transgenic plant facility at Cornell University) on the publication of this fantastic piece of work.
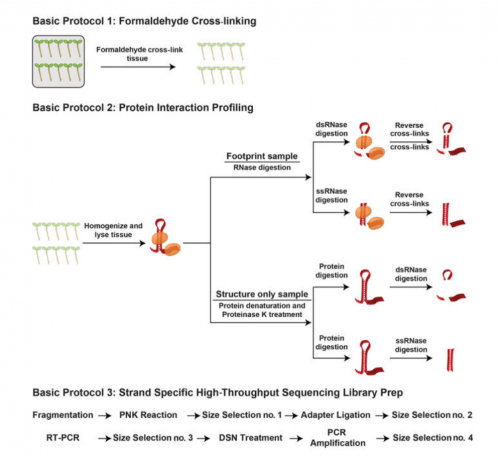
PIP-seq protocol manuscript now available
A detailed protocol for our PIP-seq approach is now available in Current Protocols in Molecular Biology. The manuscript can be found through the following link.
http://onlinelibrary.wiley.com/doi/10.1002/cpmb.21/full